Reese Lab Structure Gallery
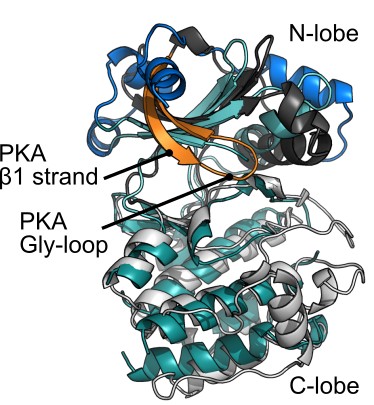
Toxoplasma With-No-Gly-Loop kinase structure
 PDB: 6M7Z
PDB: 6M7Z
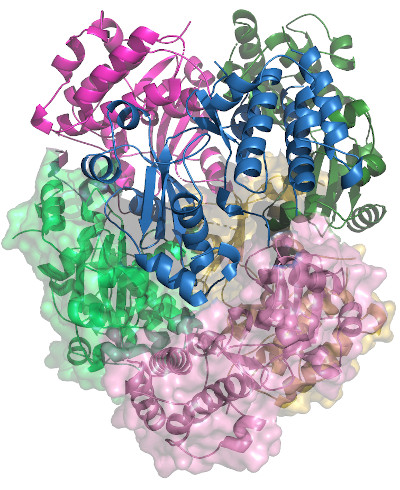
Toxoplasma ERK7 bound to the inhibitory scaffold AC9
 PDB: 6V6A
PDB: 6V6A
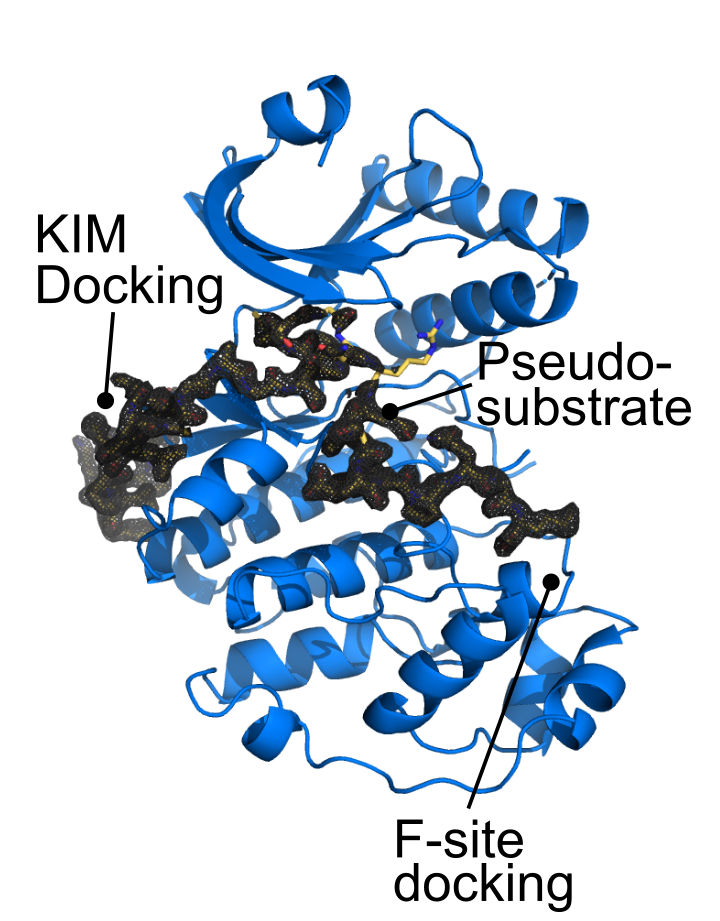
The last 30 residues of Apical cap protein 9 (AC9) wrap around the ERK7 kinase domain, making important contacts at 3 major functional sites: (1) The Kinase-interacting motif docking site, or D-site; (2) The active site; (3) the activation loop/substrate recognition site.
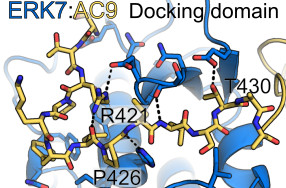
The N-terminal region region of the AC9 binding region makes contacts with conserved sites in the ERK7 D-site, though these interactions are sparse, and are insufficient to drive binding on their own.

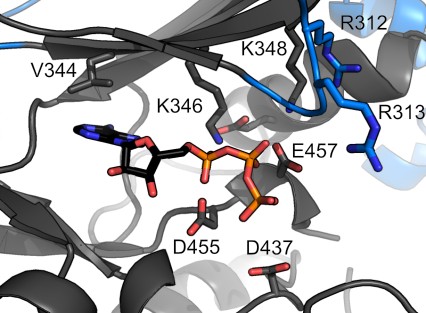
A conserved tryptophan in AC9 is buried in the ERK7 active site, acting as an ATP mimetic. The indole ring forms π-cation interactions with the catalytic Lys and hydrogen-bonds with the DFG-Asp that is normally responsible for coordinating Mg2+ ions.
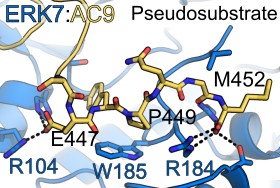
The C-terminus of AC9 contains a conserved Pro that helps anchor the protein as a pseudosubstrate. The AC9 carboxy-terminus also makes contacts with residues that normally recognize the phosphorylated ERK7 activation loop. Note that the conserved Trp in AC9 is always a set distance from the C-terminus, which would help maintain these contacts.
Conoid architecture from cryo-ET of cryo-FIB milled parasites

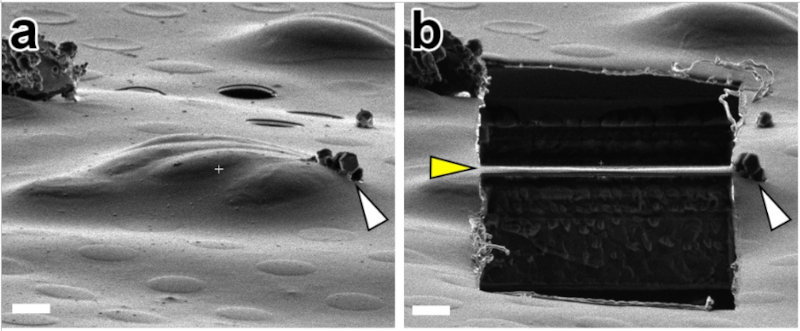
In order to visualize objects with cryo-EM or cryo-ET, they must be thin enough that the layer of water does not scatter the majority of the electrons (200 nm or less is good). At this thickness, the tension of the water can crush objects, flattening cells and their cytoskeletons. To avoid this, we used cryo-FIB milling, whereby we froze Neospora caninum parasites in "thick" ice and then milled it into a thin "lamella" using a focused beam of gallium ions.
This allowed us to get unprecedented views of the apical complex. Here is a slice of a raw tomogram of a parasite with its conoid in the retracted state, along with a segmented view in 3D. Note that this region of the parasite would be far too thick to image without FIB-milling.
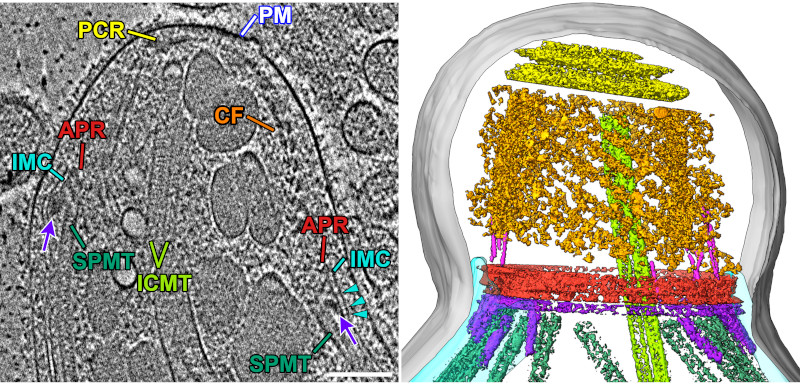
We also got great images of the protruded conoid with all membranes intact and without the structures getting squished by surface tension. Together, these data allowed us to directly compare the two states in a relatively native context.
EMDB: EMD-28247, EMD-28249, EMD-26873
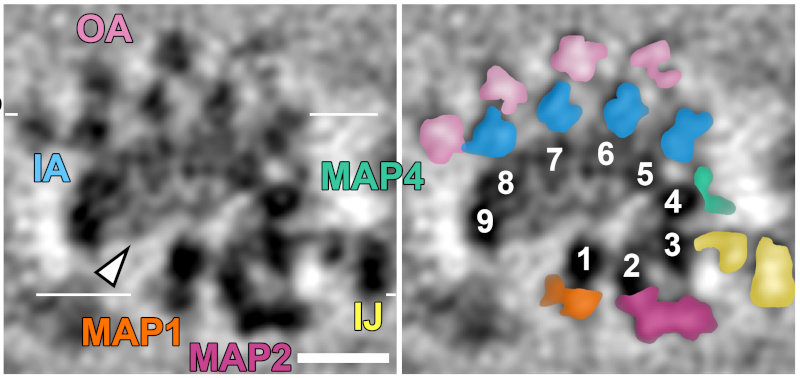
We were able to average the conoid fiber to gain a ~2 nm resolution map of the structure. In so doing we were able to clearly identify the 9 comma-shaped tubulin protofilaments that form the fiber (numbered in right panel). This is quite different from the normal tube-shape of a microtubule! We also identified density inside the fiber composed of unknown "microtubule-interacting proteins" or MIPS (arrow). Radiating out from the protofilaments is clear density for "microtubule-associated proteins" or MAPs.
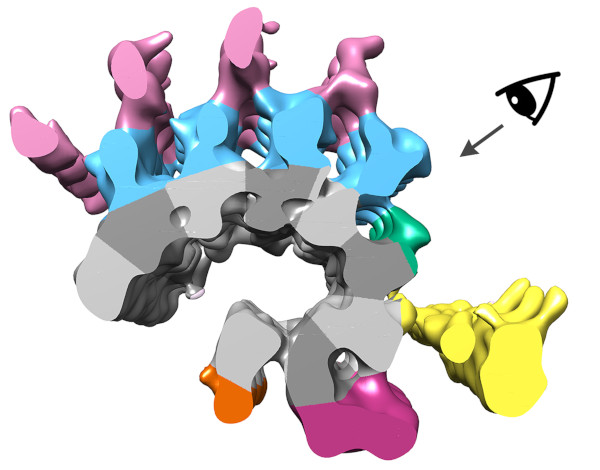
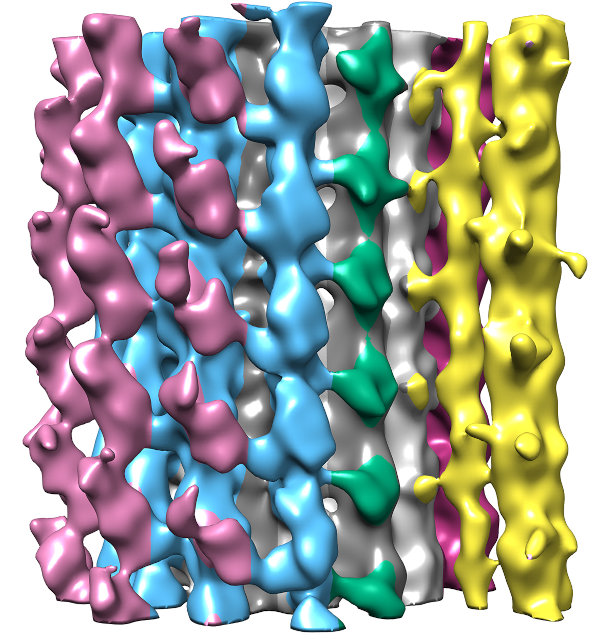
Segmenting the density from the averaged tomogram allowed us to see how these MAPs were arranged along the fiber. In fact, the MAPS appear to coat the entire surface of the fiber, which helps explain why antibodies against tubulin do not recognize the conoid fiber.
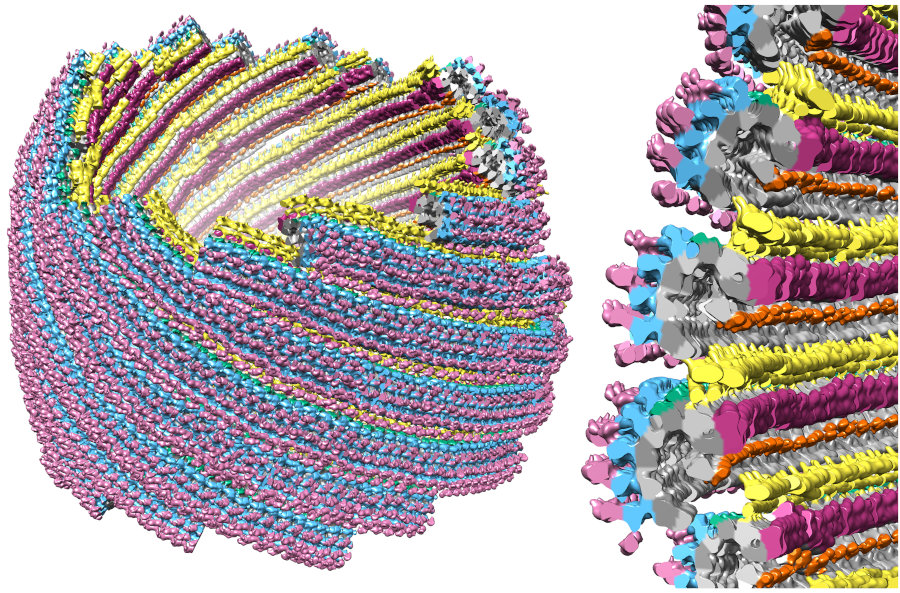
When we mapped the segmented structure back onto the full conoid, we found that these MAPs made connections between neighboring fibers on the conoid's inside surface (yellow, orange, and dark purple in image). These connections help explain why the conoid is stays rigid as it moves.
EMDB: EMD-28231, EMD-28234, EMD-28246
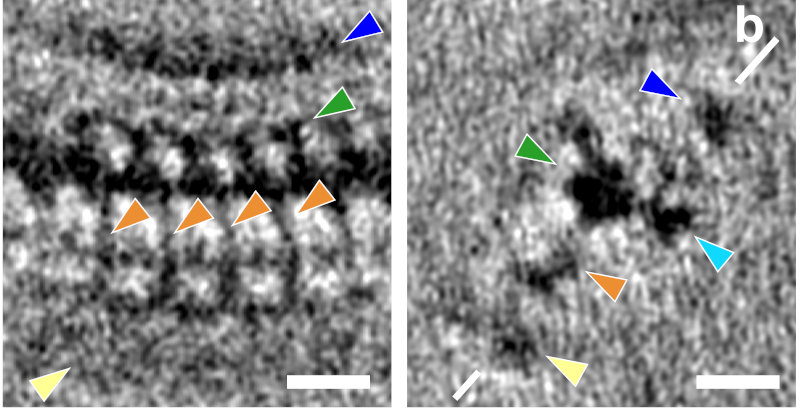
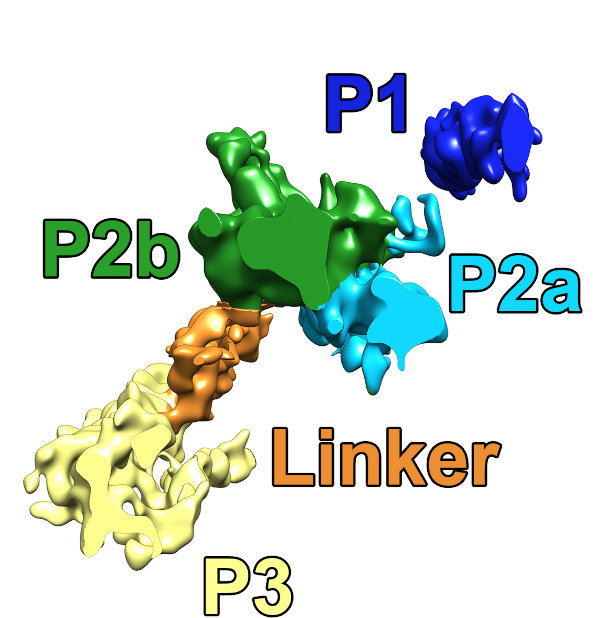
We were able to average the repeats of the pre-conoidal rings (the PCR) and found 3 major layers (P1-P3), separated by a regularly spaced linker density (orange).
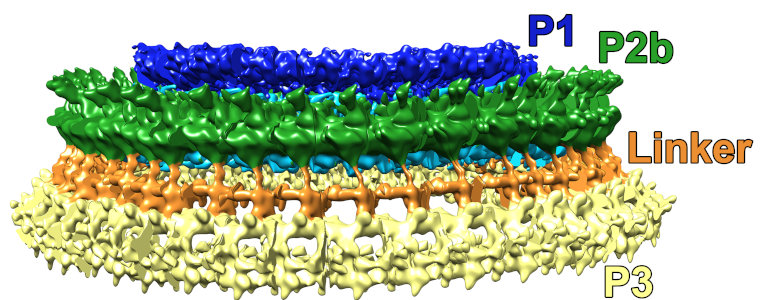
This is a side-view of the fully-segmented PCR ring. This structure is a whopping ~215 nm in diameter.
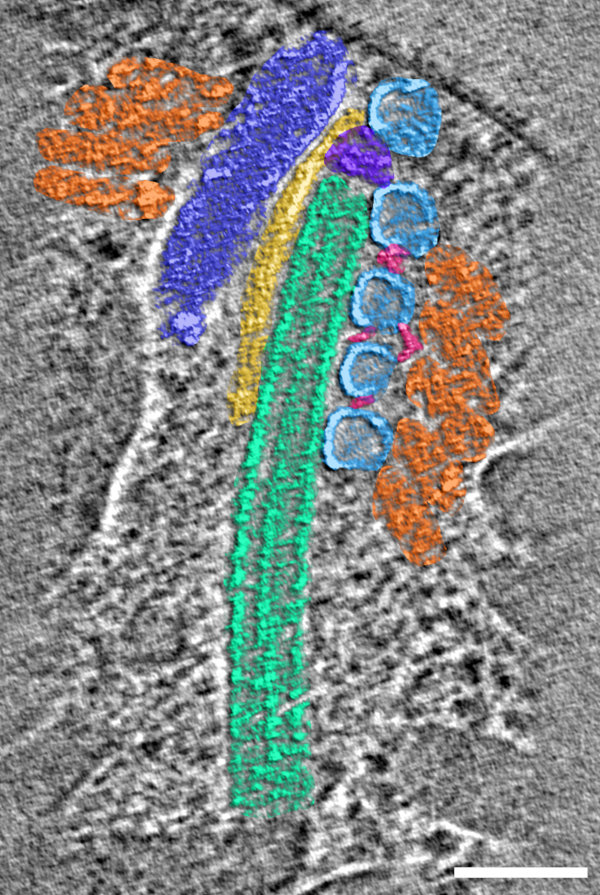
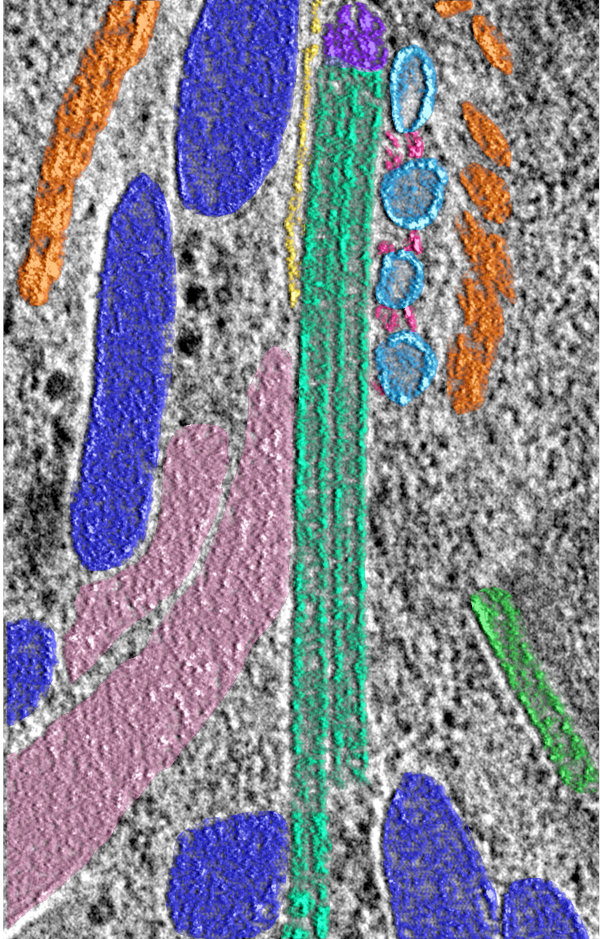
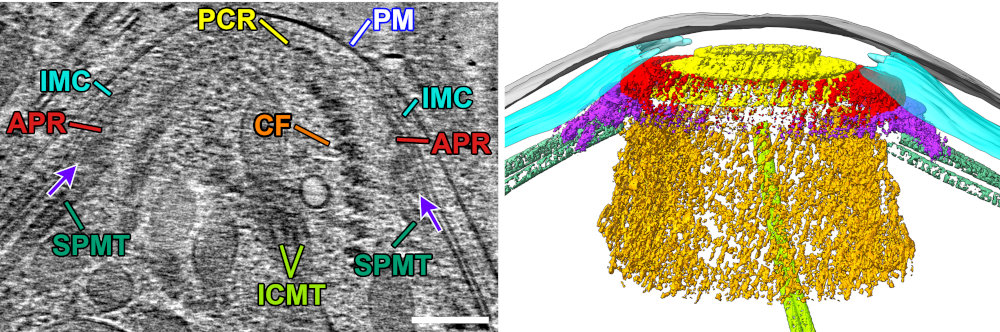
In addition to views of the cytoskeleton, we also were able to see the organization of the secretory organelles within the conoid. To the left is an annotated slice from a retracted conoid, to the right, from a protruded conoid. Colors are: conoid fibers (orange), intraconoidal microtubules (ICMT; blue-green), ICMT cap (purple), apical vesicles (light blue), vesicle connections (hot pink), micronemes (blue), rhoptries (light pink), subpellicular microtubules (lime green),
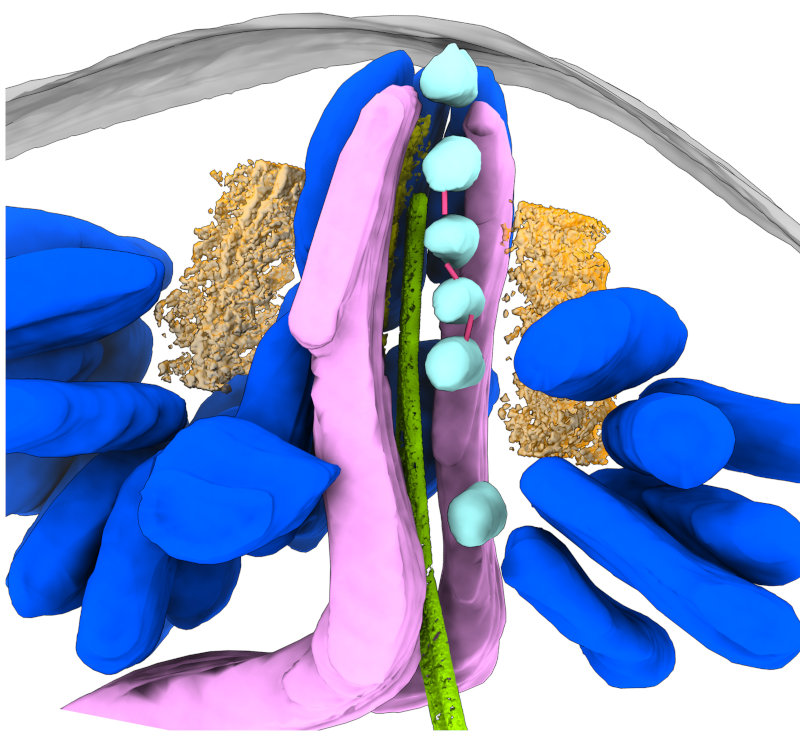
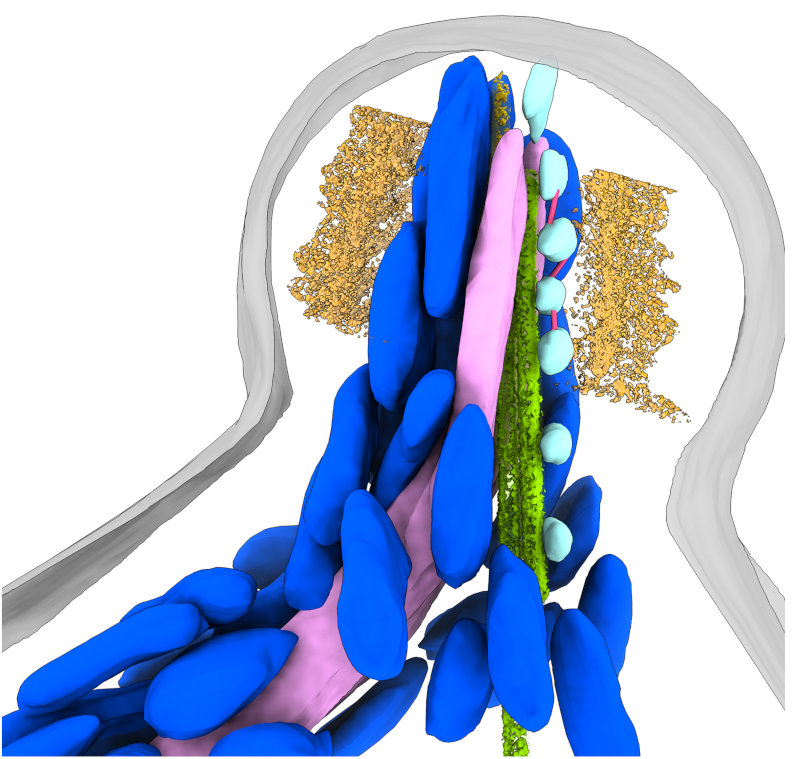
This allowed us to get a segmented overview of the secretory organelles in relationship to the conoid (orange rings in the figures) and other cytoskeletal structures (not shown). To the left is a retracted conoid, to the right is a protruded conoid.